Blog
- Home
- Blog
Learn R – Part 4

Learn R – Part 4
1. Basic Statistical Functions
R provides built-in functions for statistical analysis:
summary(): Summary statistics (min, max, quartiles, mean).
sum(): Total of values.
range(): Minimum and maximum.
var(): Variance.
sd(): Standard deviation.
Demo Data
# Basic dataset
data_basic <- c(2, 4, 6, 8, 10)
# Advanced dataset (mtcars)
data(mtcars)
mpg <- mtcars$mpg Practice
# BASIC TASKS
# HW1: Calculate the sum of data_basic
# HW2: Find the range (min and max) of data_basic
# HW3: Compute the variance of data_basic
# ADVANCED TASKS
# HW4: Calculate the standard deviation of mtcars$mpg
# HW5: Generate a summary of mtcars$hp (horsepower) Solution
# BASIC SOLUTIONS
sum_basic <- sum(data_basic)
range_basic <- range(data_basic)
var_basic <- var(data_basic)
# ADVANCED SOLUTIONS
sd_mpg <- sd(mpg)
summary_hp <- summary(mtcars$hp) 2. Mean, Median, Mode
Mean : Average (mean()).
Median : Middle value (median()).
Mode : Most frequent value (no built-in function—custom code required).
Demo Data
# Basic dataset
data_numbers <- c(1, 2, 2, 3, 4, 5, 5, 5)
# Advanced dataset (iris)
data(iris)
sepal_length <- iris$Sepal.Length Practice
# BASIC TASKS
# HW1: Calculate the mean of data_numbers
# HW2: Find the median of data_numbers
# HW3: Write a function to compute the mode
# ADVANCED TASKS
# HW4: Compute the mean of iris$Sepal.Length
# HW5: Find the median of iris$Petal.Length grouped by Species Solution
# BASIC SOLUTIONS
mean_val <- mean(data_numbers)
median_val <- median(data_numbers)
mode_func <- function(x) {
ux <- unique(x)
ux[which.max(tabulate(match(x, ux)))]
}
mode_val <- mode_func(data_numbers)
# ADVANCED SOLUTIONS
mean_sepal <- mean(sepal_length)
median_petal <- aggregate(Petal.Length ~ Species, iris, median) 3. Max, Min, and Percentiles
max()/min(): Extreme values.
quantile(): Percentiles (e.g., 25th, 50th).
IQR(): Interquartile range.
Demo Data
# Basic dataset
data_scores <- c(45, 67, 89, 34, 56, 78, 90, 23)
# Advanced dataset (airquality)
data(airquality)
temp <- airquality$Temp Practice
# BASIC TASKS
# HW1: Find the max and min of data_scores
# HW2: Calculate the 75th percentile of data_scores
# HW3: Compute the IQR of data_scores
# ADVANCED TASKS
# HW4: Find the 90th percentile of airquality$Temp
# HW5: Identify outliers in airquality$Ozone using IQR Solution
# BASIC SOLUTIONS
max_score <- max(data_scores)
min_score <- min(data_scores)
percentile_75 <- quantile(data_scores, 0.75)
iqr_score <- IQR(data_scores)
# ADVANCED SOLUTIONS
percentile_90 <- quantile(temp, 0.90)
# Outlier detection (IQR method)
q1 <- quantile(airquality$Ozone, 0.25)
q3 <- quantile(airquality$Ozone, 0.75)
iqr <- IQR(airquality$Ozone)
outliers <- airquality$Ozone[airquality$Ozone < (q1 - 1.5*iqr) | airquality$Ozone > (q3 + 1.5*iqr)] 4. Hypothesis Testing (T-Test/ANOVA)
Perform t-tests (t.test()), ANOVA (aov()), and chi-square tests (chisq.test()) to compare groups.
Demo Data
# Create sample data
group_a <- c(20, 22, 19, 18, 24)
group_b <- c(25, 24, 22, 23, 20) Practice
# HW1: Perform an independent t-test between group_a and group_b
# HW2: Run a one-way ANOVA on `mtcars` to compare `mpg` across cylinder groups Solution
# HW1
t.test(group_a, group_b)
# HW2
cyl_groups <- split(mtcars$mpg, mtcars$cyl)
anova_result <- aov(mpg ~ factor(cyl), data=mtcars)
summary(anova_result) 5. Regression Analysis (Linear Regression)
Fit linear (lm()) and logistic regression models. Use summary() to interpret coefficients and p-values.
Demo Data
# Use `mtcars` for linear regression Practice
# HW1: Fit a linear model predicting `mpg` from `wt` and `hp`
# HW2: Check the R-squared value of the model Solution
# HW1
model <- lm(mpg ~ wt + hp, data=mtcars)
# HW2
summary(model)$r.squared 6. Data Transformation
Recode variables with dplyr::mutate() and case_when(). Create new variables using arithmetic/logical operations.
Demo Data
# Create sample data
df <- data.frame(
age = c(18, 25, 30, 35, 40),
income = c(50000, 60000, 75000, 90000, 120000)
) Practice
# HW1: Recode `age` into categories: "<25", "25-35", ">35"
# HW2: Create a new variable `income_group` (Low: <70k, High: >=70k) Solution
# HW1
library(dplyr)
df <- df %>%
mutate(age_group = case_when(
age < 25 ~ "<25",
age >= 25 & age <= 35 ~ "25-35",
age > 35 ~ ">35"
))
# HW2
df <- df %>%
mutate(income_group = ifelse(income >= 70000, "High", "Low")) 7. Exporting Results
Export tables and plots using write.csv(), stargazer, or flextable.
Practice
# HW1: Save `mtcars` summary to a CSV
# HW2: Export a ggplot to PNG Solution
# HW1
write.csv(summary(mtcars), "mtcars_summary.csv")
# HW2
ggsave("plot.png", plot=last_plot()) 8. Additional Statistical Concepts
Skewness : Measure of asymmetry (moments package).
Kurtosis : Tailedness of the distribution (moments package).
Covariance : cov().
Correlation : cor().
Demo Data
# Advanced dataset (cars)
data(cars)
speed <- cars$speed
dist <- cars$dist Practice
# HW1: Calculate covariance between speed and distance
# HW2: Compute correlation between speed and distance
# HW3: Install the `moments` package and calculate skewness of speed Solution
# HW1
covariance <- cov(speed, dist)
# HW2
correlation <- cor(speed, dist)
# HW3
library(moments)
skewness_speed <- skewness(speed) 9. Handling Missing Values
Use na.rm = TRUE to ignore NA values in calculations.
Demo Data
data_missing <- c(1, 2, NA, 4, 5) Practice
# HW1: Calculate the mean of data_missing (ignore NA)
# HW2: Check if data_missing contains any NA values Solution
mean_missing <- mean(data_missing, na.rm = TRUE)
has_na <- anyNA(data_missing) Learn R – Part 3

Learn R – Part 3
1. Plot (Data Visualization)
Plots visualize trends and relationships. Use plot() for basic graphs, lines()/points() for overlays, and par() for layouts. Customize with main, xlab, ylab, col, lwd, pch, and bg.
Demo Data (Basic)
# Simple dataset for beginners
x <- 1:10
y <- c(2, 4, 6, 8, 7, 5, 3, 1, 9, 10) Demo Data (Advanced)
# Complex dataset: mtcars (built-in)
data(mtcars)
mpg <- mtcars$mpg # Miles per gallon
hp <- mtcars$hp # Horsepower
wt <- mtcars$wt # Weight Practice
# BASIC TASKS
# HW1: Plot x vs. y as points
# HW2: Add a blue line to the plot
# HW3: Create a plot with red triangles (pch=17)
# ADVANCED TASKS
# HW4: Plot mpg vs. hp from mtcars, add a smooth line
# HW5: Create a multi-plot layout (2x2 grid)
# HW6: Customize mpg vs. wt plot: title, axis labels, green points, gray background Solution
# BASIC SOLUTIONS
plot(x, y)
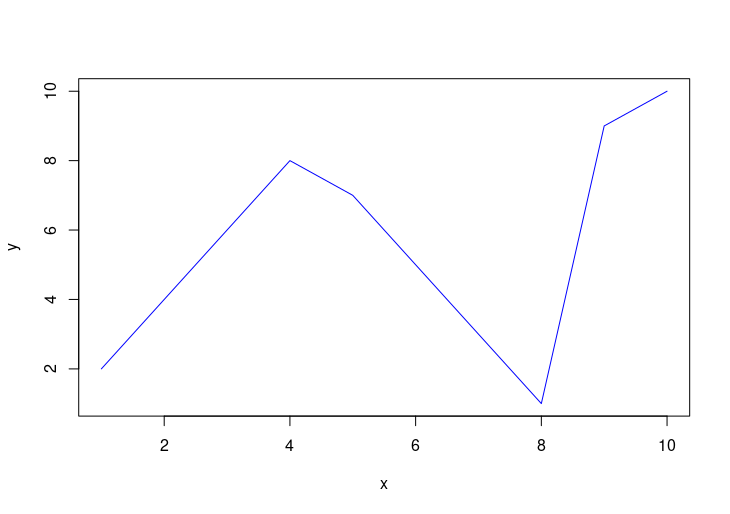
plot(x, y, type="l", col="blue")
plot(x, y, pch=17, col="red")
# ADVANCED SOLUTIONS
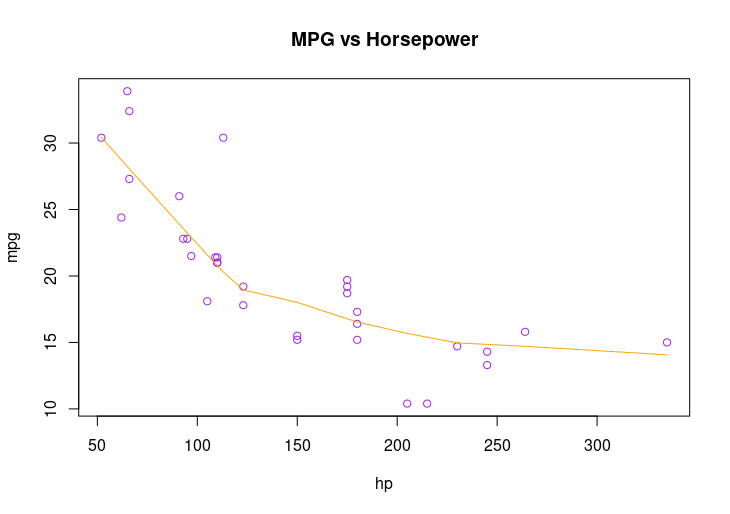
plot(mpg ~ hp, data=mtcars, main="MPG vs Horsepower", col="purple")
lines(lowess(hp, mpg), col="orange")
par(mfrow=c(2,2))
plot(mpg ~ hp, data=mtcars)
plot(mpg ~ wt, data=mtcars)
hist(mpg, col="lightblue")
boxplot(mpg, col="yellow")
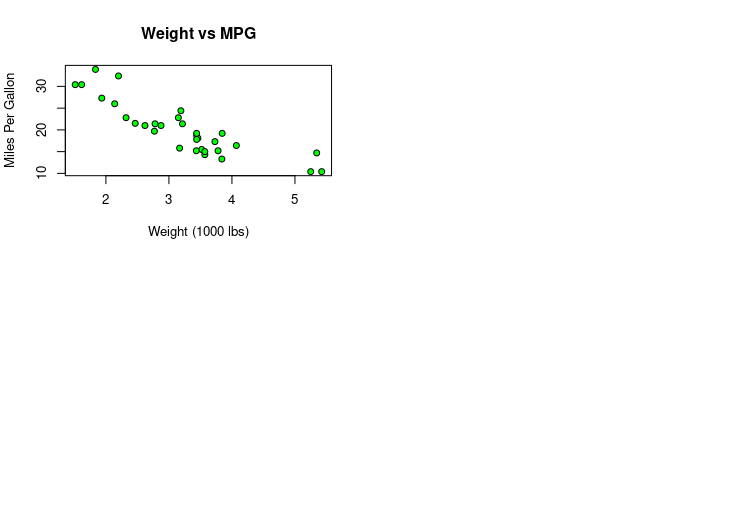
plot(mpg ~ wt, data=mtcars,
main="Weight vs MPG",
xlab="Weight (1000 lbs)",
ylab="Miles Per Gallon",
pch=21, bg="green",
panel.first=grid()) 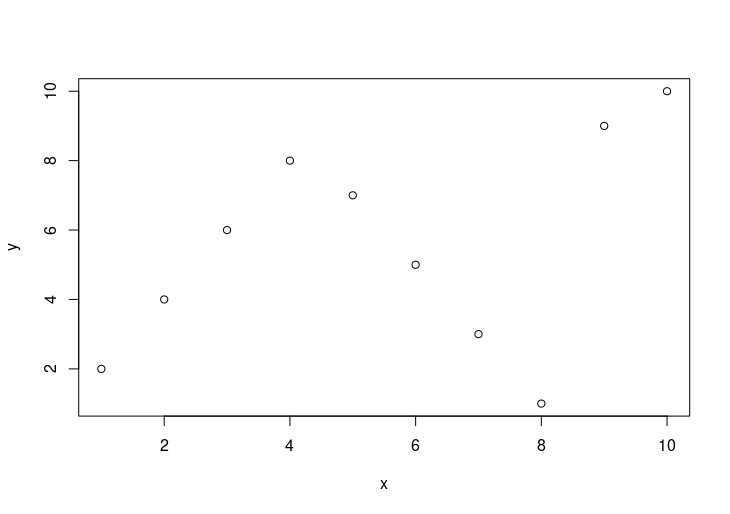
HW1: Plotting of (x, y)
HW2: Adding a blue line to the plot

HW3: A plot with red triangles (pch=17)
HW4: Plot mpg vs. hp from mtcars, add a smooth line
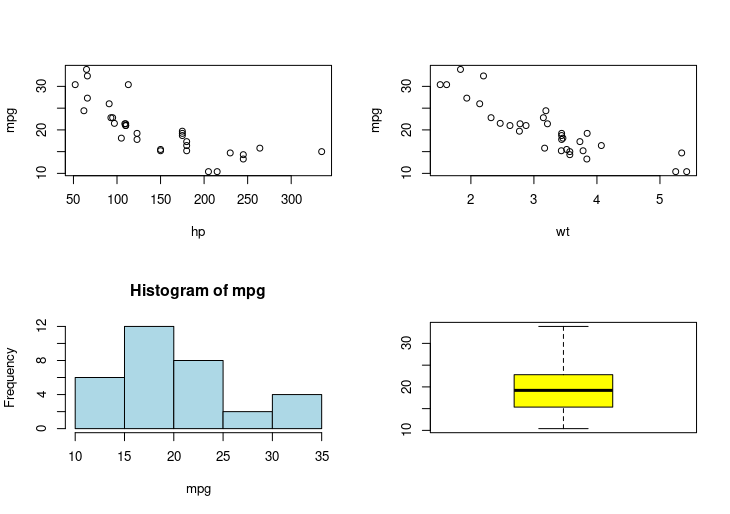
HW5: A multi-plot layout (2×2 grid)
HW6: Customization mpg vs. wt plot
2. Pie Chart
Pie charts show proportions. Use pie(), legend(), and ifelse() for conditional formatting.
Demo Data (Basic)
# Simple sales data
sales <- c(25, 35, 40)
labels <- c("Apparel", "Electronics", "Groceries") Demo Data (Advanced)
# Complex dataset: Titanic survival rates
survivors <- c(203, 118, 178, 528)
groups <- c("1st Class", "2nd Class", "3rd Class", "Crew") Practice
# BASIC TASKS
# HW1: Create a pie chart for sales data
# HW2: Add a title and explode the "Groceries" slice
# ADVANCED TASKS
# HW3: Plot Titanic survival rates with gradient colors
# HW4: Add a legend and percentage labels
# HW5: Create a 3D pie chart (use plotrix package) Solution
# BASIC SOLUTIONS
pie(sales, labels=labels)
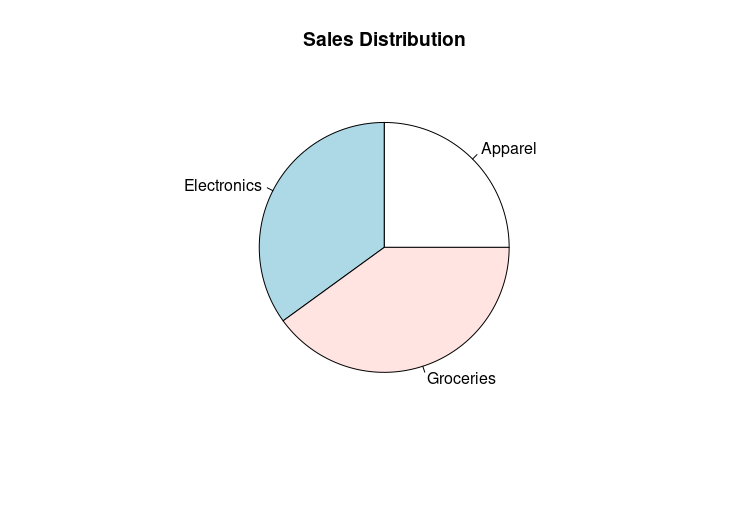
pie(sales, labels=labels, main="Sales Distribution", explode=0.1)
# ADVANCED SOLUTIONS
# Install the plotrix package (only needed once)
install.packages("plotrix")
# Load the package
library(plotrix)
library(plotrix)
pie3D(survivors, labels=groups, main="Titanic Survival Rates",
col=heat.colors(4), explode=0.05)
pie(survivors, labels=paste(groups, " (", round(survivors/sum(survivors)*100, 1), "%)", sep=""),
col=rainbow(4))
legend("right", groups, fill=rainbow(4)) 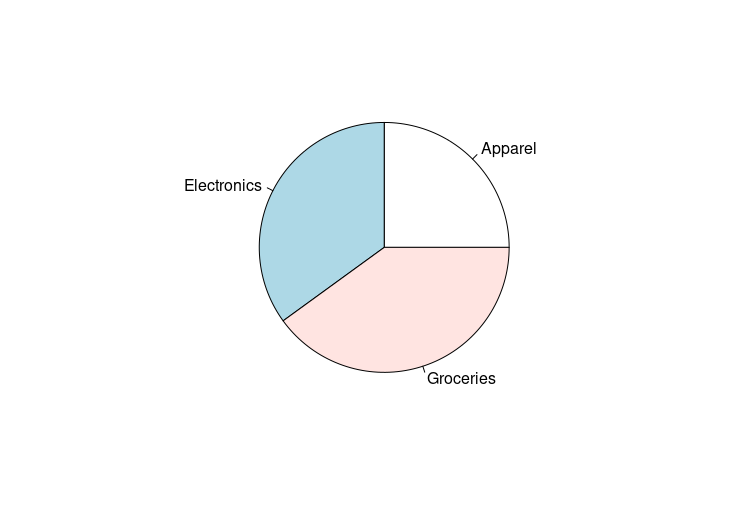
HW1: A pie chart for sales data
HW2: Adding a title and explode the “Groceries” slice
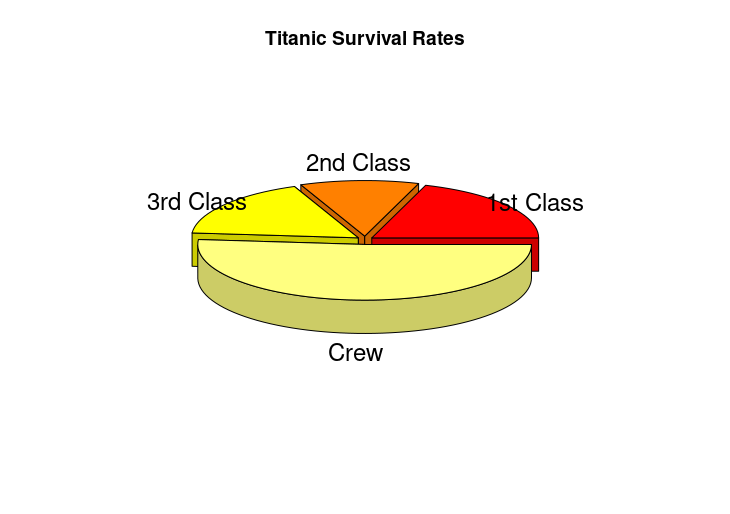
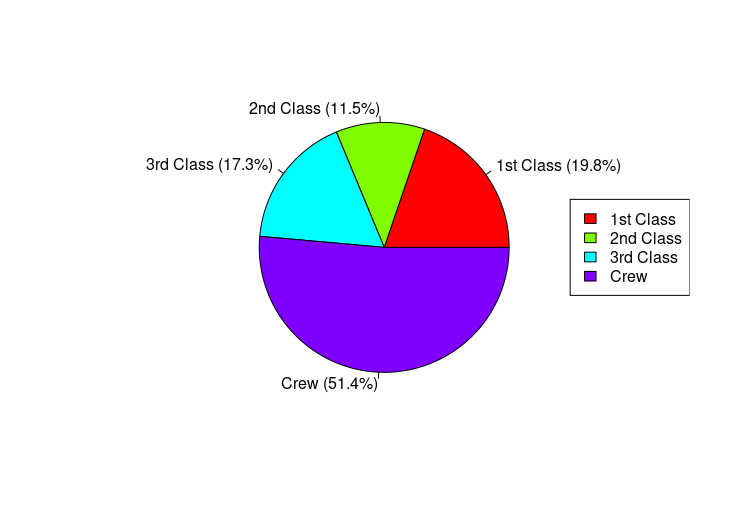
HW3: Titanic survival rates with gradient colors
HW4: Legend and percentage labels
3. Bar Chart
Bar charts compare categories. Use barplot(), beside=TRUE for grouped bars, and col for gradients.
Demo Data (Basic)
# Monthly sales
months <- c("Jan", "Feb", "Mar")
sales <- c(200, 450, 300) Demo Data (Advanced)
# Complex dataset: Olympic medal counts
countries <- c("USA", "China", "Russia", "UK")
gold <- c(39, 38, 20, 22)
silver <- c(41, 32, 28, 21)
bronze <- c(33, 18, 23, 22) Practice
# BASIC TASKS
# HW1: Create a vertical bar chart for monthly sales
# HW2: Add grid lines and rotate labels
# ADVANCED TASKS
# HW3: Create stacked bars for Olympic medals
# HW4: Create grouped bars with legends
# HW5: Add error bars using arrows() Solution
# BASIC SOLUTIONS
barplot(sales, names.arg=months, main="Monthly Sales", xlab="Month", ylab="Revenue")
barplot(sales, names.arg=months, las=2, cex.names=0.8, col="lightgreen")
abline(h=seq(0, 500, by=100), lty=2)
# ADVANCED SOLUTIONS
medals <- rbind(gold, silver, bronze)
barplot(medals, names.arg=countries, col=c("gold", "silver", "darkorange"),
legend=rownames(medals), main="Olympic Medals", beside=TRUE)
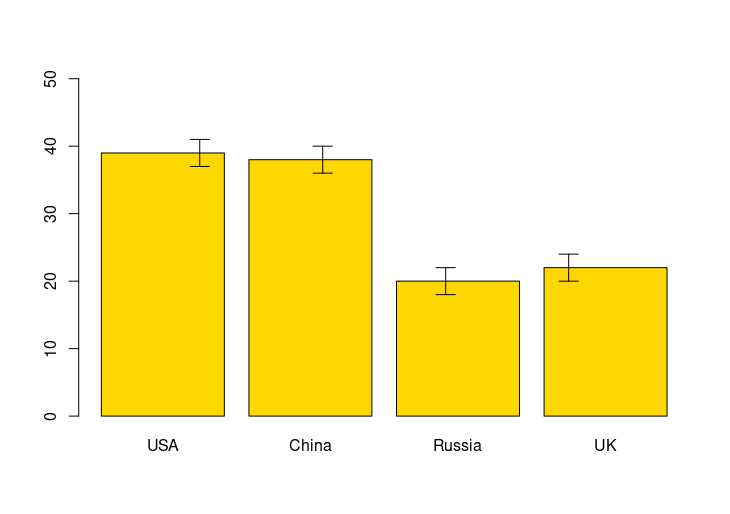
# Error bars
barplot(gold, names.arg=countries, ylim=c(0, 50), col="gold")
arrows(x0=1:4, y0=gold-2, y1=gold+2, code=3, angle=90, length=0.1) 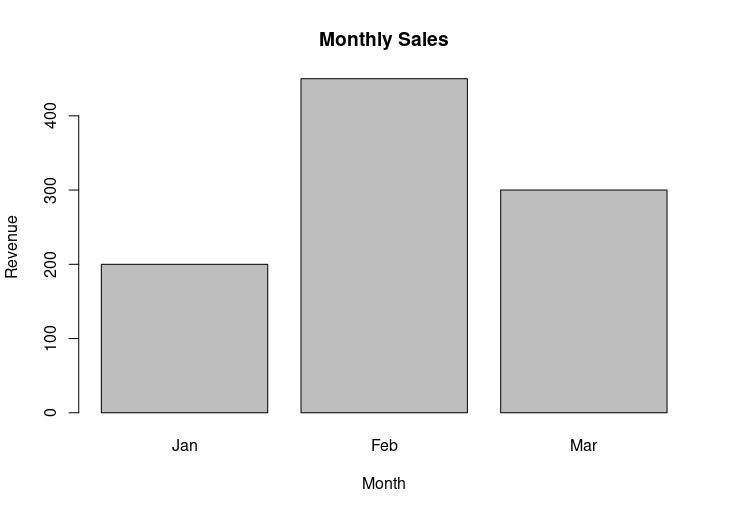
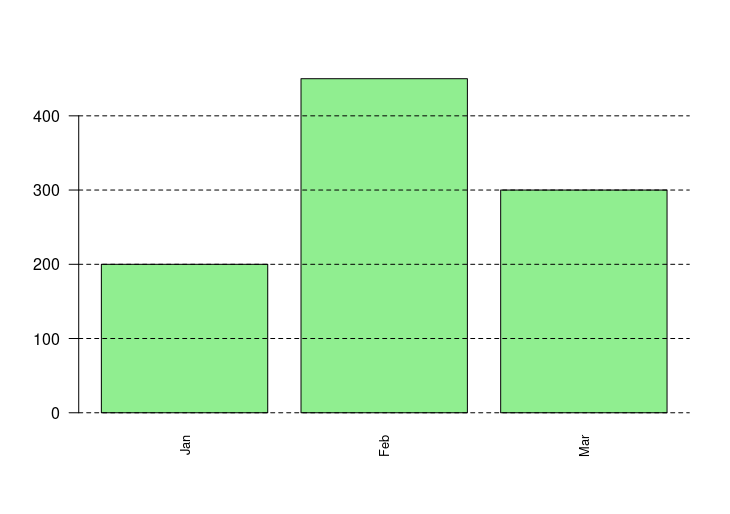
HW1: A vertical bar chart for monthly sales
HW2: Adding grid lines and rotate labels
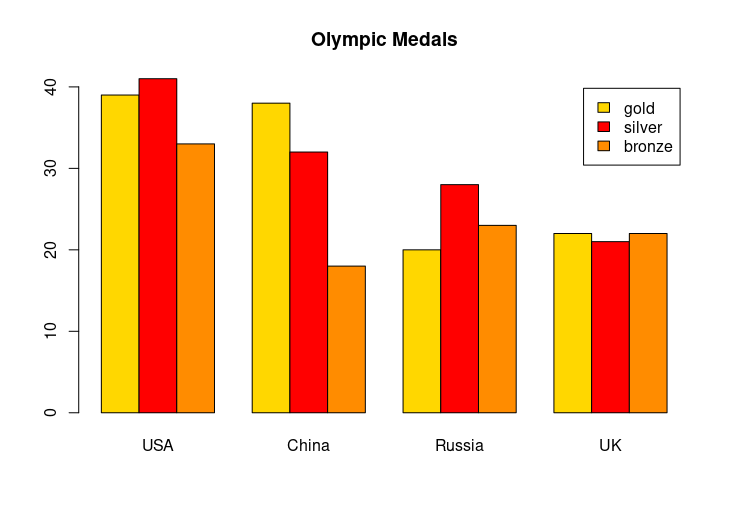
HW3: Stacked bars for Olympic medals
HW4: Add error bars using arrows()
Learn R – Part 2

Learn R – Part 2
1. Vectors
Vectors are 1D data structures holding elements of the same type .
Sort : sort() (ascending) or rev(sort()) (descending).
Create : Use c(), seq(from, to, by), or rep(value, times).
Access : Use [index] (positive/negative), logical vectors, or names.
Length : length(vector).
# HW1: Create a vector of even numbers 2, 4, 6 using `seq()`
# HW2: Access the 3rd element of `c(10, 20, 30, 40)`
# HW3: Sort `c(5, 1, 3)` in descending order
# HW4: Check the length of `c("a", "b", "c")`
# HW5: Create a vector with 3 copies of "R" using `rep()` # HW1
vec_seq <- seq(2, 6, by=2) # Output: 2, 4, 6
# HW2
print(c(10, 20, 30, 40)[3]) # Output: 30
# HW3
sorted <- rev(sort(c(5, 1, 3))) # Output: 5, 3, 1
# HW4
print(length(c("a", "b", "c"))) # Output: 3
# HW5
vec_rep <- rep("R", 3) # Output: "R", "R", "R" 2. Lists
Lists store mixed or nested data .
Add/Remove : list[[new_index]] <- value or list[index] <- NULL.
Create : list().
Access : [index] (returns sublist), [[index]] (returns element), or $name.
Modify : Assign new values via [[ ]] or append().
# HW1: Create a list with "apple", 25, and a sub-list `c(1, 2)`
# HW2: Access the sub-list `c(1, 2)` from HW1
# HW3: Change "apple" to "banana" in the list
# HW4: Add `TRUE` to the end of the list
# HW5: Remove the 2nd element (25) # HW1
my_list <- list("apple", 25, list(1, 2))
# HW2
print(my_list[[3]]) # Output: 1, 2
# HW3
my_list[[1]] <- "banana"
# HW4
my_list <- append(my_list, TRUE)
# HW5
my_list[2] <- NULL 3. Matrices
Matrices are 2D, same-type data structures.
Generate Values : seq(), rep().
Create : matrix(data, nrow, ncol).
Access : [row, col], [,] for entire rows/columns.
Add Rows/Columns : rbind(), cbind().
# HW1: Create a 3x2 matrix with 1-6 using `matrix()`
# HW2: Extract the 2nd row
# HW3: Extract the 1st column
# HW4: Add a row `7, 8` to the matrix
# HW5: Create a matrix with 1, 2 repeated 3 times using `rep()` # HW1
mat <- matrix(1:6, nrow=3)
# HW2
print(mat[2, ]) # Output: 2, 5
# HW3
print(mat[, 1]) # Output: 1, 2, 3
# HW4
mat <- rbind(mat, c(7, 8))
# HW5
mat_rep <- matrix(rep(1:2, 3), nrow=3) 4. Arrays
Arrays extend matrices to multi-dimensional data .
Dimensions : dim() to check or set dimensions.
Create : array(data, dim=c(rows, cols, ...)).
Access : [i, j, k] for specific elements.
# HW1: Create a 2x2x2 array with values 1-8
# HW2: Access the 3rd element of the 1st layer
# HW3: Extract the 2nd layer (all rows/columns)
# HW4: Check the total length of the array
# HW5: Convert a vector `1:12` into a 3x4 array # HW1
arr <- array(1:8, dim=c(2, 2, 2))
# HW2
print(arr[3]) # Output: 3
# HW3
print(arr[, , 2])
# HW4
print(length(arr)) # Output: 8
# HW5
arr_3d <- array(1:12, dim=c(3, 4)) 5. Data Frames
Data frames store tabular data (mixed types allowed).
Modify : Add/remove columns via $ or [ ].
Create : data.frame().
Access : $column, [, "column"], or subset().
# HW1: Create a data frame with Name (Alice, Bob), Age (25, 30)
# HW2: Access the "Name" column using `$`
# HW3: Add a column "Salary" with 5000, 6000
# HW4: Remove the "Age" column
# HW5: Check the number of rows # HW1
df <- data.frame(Name=c("Alice", "Bob"), Age=c(25, 30))
# HW2
print(df$Name) # Output: Alice, Bob
# HW3
df$Salary <- c(5000, 6000)
# HW4
df$Age <- NULL
# HW5
print(nrow(df)) # Output: 2 6. Factors
Factors store categorical data with predefined levels.
Ordered Factors : ordered=TRUE for ranking.
Create : factor().
Modify Levels : levels(), factor(..., levels=).
# HW1: Create a factor with "Low", "Medium", "High"
# HW2: Check the levels of the factor
# HW3: Add "Very High" as a new level
# HW4: Remove "Medium" from the factor
# HW5: Convert the factor to an ordered factor # HW1
f <- factor(c("Low", "Medium", "High"))
# HW2
print(levels(f)) # Output: "High", "Low", "Medium"
# HW3
f <- factor(f, levels=c("Low", "Medium", "High", "Very High"))
# HW4
f <- f[f != "Medium"]
# HW5
f_ordered <- factor(f, ordered=TRUE) Learn R – Part 1

Learn R – Part 1
1. How to Write an R Code (Syntax and Print)
R uses simple syntax to execute commands. Code is written line-by-line, and print() or cat() displays output. Try, if you can solve the below problem.
# HW 1: Print "Hello, R!"
# HW 2: Calculate 5 + 3 and print the result print("Hello, R!")
cat("Result:", 5 + 3) 2. To See a Data Type
Use class() / typeof() to check the data type of a variable (e.g., numeric, character).
# HW 1: Check the type of "apple"
# HW 2: Check the type of 25.6 class("apple") # Output: "character"
typeof(25.6) # Output: "double" 3. To Set a Data Type
Use as. function to set forcefully a data type like as.numeric() or as.character().
# HW 1: Convert "100" to numeric
# HW 2: Convert 50 to character as.numeric("100") # Output: 100
as.character(50) # Output: "50" 4. To Handle Multiple values at once
Use vectors (created with c()) to store multiple values of the same type.
# HW 1: Create a vector of numbers 1, 2, 3
# HW 2: Create a vector of colors: "red", "blue" nums <- c(1, 2, 3)
colors <- c("red", "blue") 5. To Define a Value
Assign values to variables using <- or =. Example: x <- 10.
# HW 1: Assign 3.14 to "pi"
# HW 2: Assign "Rocks" to "language" pi <- 3.14
language <- "Rocks" 6. How to make a Comment
Use # to add comments (notes) to your code. R ignores these lines.
# HW 1: Write a comment explaining the next line
print("Learning R!") # This line prints a motivational message
print("Learning R!") 7. Variables and Their Types
Variables in R store data, and their type is automatically inferred from the assigned value. Use <- or = to assign values. Example: x <- 10 (numeric), y <- "Hello" (character).
# HW1: Assign 3.14 to a variable
# HW2: Assign "R Programming" to a variable
# HW3: Assign TRUE to a variable pi_value <- 3.14
language <- "R Programming"
is_fun <- TRUE 8. Different Data Types in R
R supports numeric (e.g., 10.5), integer (e.g., 55L), complex (e.g., 9+3i), character (e.g., "R is exciting"), and logical (e.g., TRUE). Use class() to check the type.
# HW1: Create a numeric variable with 787
# HW2: Create an integer with 100L
# HW3: Create a complex number 9+3i
# HW4: Create a character "FALSE"
# HW5: Create a logical FALSE num <- 787
integer_val <- 100L
complex_num <- 9 + 3i
char <- "FALSE"
logical_val <- FALSE 9. Simple Math Functions
R has built-in functions for math operations:
min(): Finds the smallest value.
floor(): Rounds down to the nearest integer.
max(): Finds the largest value.
sqrt(): Calculates the square root.
abs(): Returns absolute value.
ceiling(): Rounds up to the nearest integer.
# HW1: Find the minimum of 10, 20, 5
# HW2: Calculate the square root of 16
# HW3: Round 3.2 up and 4.9 down
# HW4: Get the absolute value of -7.5 min_val <- min(10, 20, 5) # Output: 5
sqrt_val <- sqrt(16) # Output: 4
ceil_val <- ceiling(3.2) # Output: 4
floor_val <- floor(4.9) # Output: 4
abs_val <- abs(-7.5) # Output: 7.5 10. Boolean’s (Logical Values)
Booleans are TRUE or FALSE. Use logical operators like & (AND), | (OR), and ! (NOT) to combine conditions.
# HW1: Check if 5 > 3 AND 2 < 4
# HW2: Check if 10 == 10 OR 7 != 7
# HW3: Reverse the value of TRUE bool1 <- (5 > 3) & (2 < 4)
bool1 # Output: TRUE
bool2 <- (10 == 10) | (7 != 7)
bool2 # Output: TRUE
bool3 <- !TRUE
bool3 # Output: FALSE
11. Arithmetic Operators
Used for basic math operations: + (add), - (subtract), * (multiply), / (divide), ^ (exponent), %% (modulus).
# HW1: Calculate 10 + 5
# HW2: Calculate 10 - 5
# HW3: Calculate 4 * 6
# HW4: Calculate 20 / 4
# HW5: Calculate 3^2 (3 squared)
# HW6: Find the remainder of 10 %% 3 add <- 10 + 5 # 15
sub <- 10 - 5 # 5
mul <- 4 * 6 # 24
div <- 20 / 4 # 5
exp <- 3^2 # 9
mod <- 10 %% 3 # 1 12. Assignment Operators
Assign values to variables: <-, =, or -> (rarely used). Example: x <- 5 or 5 -> x.
# HW1: Assign 10 to `a` using `<-`
# HW2: Assign 20 to `b` using `=`
# HW3: Assign 30 to `c` using `->` a <- 10
b = 20
30 -> c 13. Comparison Operators
Compare values: == (equal), != (not equal), >, <, >=, <=. Returns TRUE or FALSE.
# HW1: Check if 5 == 5
# HW2: Check if 10 != 5
# HW3: Check if 8 > 3
# HW4: Check if 4 <= 4 5 == 5 # TRUE
10 != 5 # TRUE
8 > 3 # TRUE
4 <= 4 # TRUE 14. Logical Operators
Combine conditions: & (AND), | (OR), ! (NOT).
# HW1: Check if (5 > 3) & (2 < 4)
# HW2: Check if (10 == 5) | (3 != 3)
# HW3: Reverse the result of TRUE using `!` (5 > 3) & (2 < 4) # TRUE
(10 == 5) | (3 != 3) # FALSE
not_op <- !TRUE
not_op # FALSE 15. If-Else Statements
Use if to execute code if a condition is TRUE, and else for when it’s FALSE.
# HW1: Check if 10 is greater than 5, print "Yes"
# HW2: Check if 7 is even; if not, print "Odd"
# HW3: Assign "Pass" if score >= 50, else "Fail" # HW1
if (10 > 5) {
print("Yes")
}
# HW2
if (7 %% 2 == 0) {
print("Even")
} else {
print("Odd")
}
# HW3
score <- 55
result <- if (score >= 50) "Pass" else "Fail"
print(result) 16. While Loop
Repeats code while a condition is TRUE. Use break to exit early.
# HW1: Print numbers 1 to 5
# HW2: Sum numbers from 1 to 3 # HW1
i <- 1
while (i <= 5) {
print(i)
i <- i + 1
}
# HW2
total <- 0
j <- 1
while (j <= 3) {
total <- total + j
j <- j + 1
}
print(total) # Output: 6 17. For Loop
Iterates over a sequence (like a vector). Use break/next to control flow.
# HW1: Print numbers 1 to 3 from c(1, 2, 3)
# HW2: Calculate sum of 1, 2, 3 using a loop
# HW3: Loop through a matrix (2x2) and print values # HW1
for (num in c(1, 2, 3)) {
print(num)
}
# HW2
sum <- 0
for (i in 1:3) {
sum <- sum + i
}
print(sum) # Output: 6
# HW3
mat <- matrix(1:4, nrow=2)
for (row in 1:nrow(mat)) {
for (col in 1:ncol(mat)) {
print(mat[row, col])
}
} 18. Functions
Reusable code blocks. Use function() to define, and return() to output results.
# HW1: Create a function to square a number
# HW2: Create a function that says "Hello, [name]!" # HW1
square <- function(x) {
return(x^2)
}
print(square(4)) # Output: 16
# HW2
greet <- function(name) {
return(paste("Hello,", name, "!"))
}
print(greet("Alice")) # Output: "Hello, Alice!" 
Course Review: How to Write and Publish a Scientific Paper
Recently, I completed the Coursera course “How to Write and Publish a Scientific Paper.” As someone new to research, I found this course incredibly valuable for building a strong foundation. It breaks down the research process step by step, from understanding the basics to confidently preparing your work for publication.
The course covers essential topics like structuring your paper, choosing the right journal, and navigating the peer-review process. What stood out most to me was how beginner-friendly yet comprehensive the lessons were—perfect for anyone starting their research journey.
If you’re looking to demystify academic writing and take your first step into the world of publishing, I highly recommend this course.
Course Overview
This project-centered course is meticulously designed to lead students through the entire process of writing and publishing a scientific paper. It encompasses:
- Understanding Academia: Provides insights into the academic publishing landscape, including how scientific journals operate and the ethical considerations involved.
- Delimiting Your Scientific Paper: Guides on defining the scope and structure of your paper, ensuring clarity and focus.
- Writing the Paper: Offers practical advice on crafting each section of the manuscript, from the introduction to the conclusion, and emphasizes the importance of proper citation practices.
- Post-Writing Checklist: Equips students with a comprehensive checklist to self-assess their work before submission, enhancing the likelihood of acceptance.
Personal Experience
As a novice in the research domain, I found this course to be exceptionally beneficial. The step-by-step approach demystified the complexities of scientific writing. The emphasis on ethical considerations and the peer-review process provided a holistic understanding of academic publishing. The practical assignments, particularly outlining a complete scientific paper and selecting an appropriate journal for submission, were instrumental in reinforcing the concepts learned.
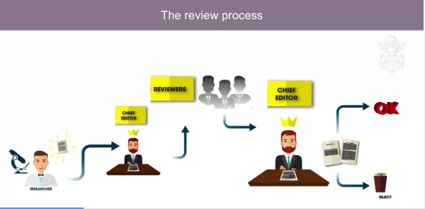
Here are some interesting screenshots of the course that you might find interesting ;
Screenshot Credit: Coursera
For any kind of Query or problem feel free to contact me in my Email